>> Download CGBayesNets <<CGBayesNetsby Michael McGeachie and Hsun-Hsien Chang.
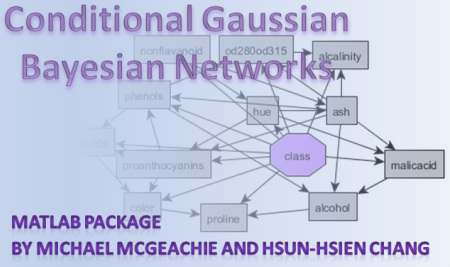
CGBayesNets builds and predicts with conditional Gaussian Bayesian networks (CGBNs), enabling biological researchers to infer predictive networks based on multimodal genomic datasets. The package provides many other functions for supporting all phases of model exploration and verification, including cross validation, bootstrapping, and AUC manipulation. CGBayesNets now comes integrated with three useful network learning algorithms : K2, Pheno-Centric, and a Full-Exhaustive greedy search. K2 is a traditional bayesian network learning algorithm that is appropriate for building networks that prioritize a particular phenotype for prediction; but it is not guaranteed to maximize prediction. A Pheno-centric network can be built around a discrete variable that maximizes predictive accuracy of the network to predict the phenotype node. The Full-Exhaustive method can be used on domains of limited size to provide a complete picture of the strongest statistical interactions among variables without biasing the network toward phenotype prediction. In all cases, we use true Bayesian inference : in network building to maximize the posterior likelihood of the data given the model using the Bayesian lilkelihood formulation of Ramoni and Chang[1]; and in inference we implement algorithm of Cowell[2] to compute probabilities of assignments to the prediction variable. Conditional Gaussian Bayesian Networks were first described by Heckerman and Geiger[3], and are a modeling technique that combines discrete and continuous variables into a Bayesian Network, where typical Bayesian Networks are limited to discrete variables only. This represents an important distinction between CGBayesNets and other free Bayesian network software. This package is available as Open Source software. Some familiarity with MATLAB is required. For more information, email mmcgeach (at) csail (dot) mit (dot) edu, or fill in the form below. If you use the CGBayesNets Package in your research, please cite the following reference: McGeachie MJ, Chang HH, Weiss ST. CGBayesNets: Conditional Gaussian Bayesian Network Learning and Inference with Mixed Discrete and Continuous Data. PLoS Computational Biology. 2014 Jun 12;10(6):e1003676. doi: 10.1371/journal.pcbi.1003676. eCollection 2014. Refs: [1]Chang HH, Ramoni MF, "Robust cross-race gene expression analysis," Proc. IEEE International Conference Acoustics, Speech, and Signal Processing 2009 (ICASSP '09), Taipei, Taiwan, April 19-24, 2009, pp. 505-508. [2]Cowell RG. Local Propagation in Conditional Gaussian Bayesian Networks. Machine Learninig Research 2005;6:1517-50. [3]Heckerman D, Gieger D. Learning Bayesian Networks: A unification for discrete and Gaussian domains. In: Uncertainty in Artificial Intelligence: Morgan Kaufmann; 1995. Contact
mmcgeach at csail dot mit dot edu
|
CGBayesNets Software Article in PLoS Computational Biology :
|